- aDNA
- Traditional Sequencing
- Next Gen Sequencing
- Chromosomes
- Data Files
PhaDNA lab
Ancient or palaeo DNA refers to genetic material obtained from old specimens. Typically, DNA degrades rapidly in dead organisms, but trace amounts are preserved in various tissues either naturally (e.g. unmineralised bird bones or faeces in dry caves) or artificially (e.g. early museum specimens collected for morphology). Access to palaeoDNA provides many opportunities for research by allowing inclusion of rare or extinct species or populations, matching taxonomic material to extant populations and sampling population genetics through time etc. A critical aspect of palaeoDNA studies is the avoidance of contamination from modern DNA and in particular amplified DNA (via PCR).
The Phoenix AncientDNA lab is a dedicated facility for the preparation and handling of palaeoDNA material from fossil, herbarium and museum samples. PhaDNA is located in a building remote from neoDNA labs associated with the research group. Access to the lab is strictly controlled and limited requiring maglock swipe card authorisation.
All users are required to follow strict protocols that preclude use of neoDNA facilities prior to the same day session. No PCR products or modern samples are used in or near the PhaDNA lab. The purpose-built lab is equipped with air-lock/gown-up space, positive pressure, UV decontamination and filtered-air PCR setup. Dedicated equipment includes refrigerator, freezer, 3 benchtop centrifuges, vortex, HEPA-filtered PCR setup hood, fume hood, hybridization ovens, telephone and networked computer etc. The lab meets current safety specifications.
Current research using the lab includes studies of insect population genetics and bird phylogenetics. |

|
  |

|
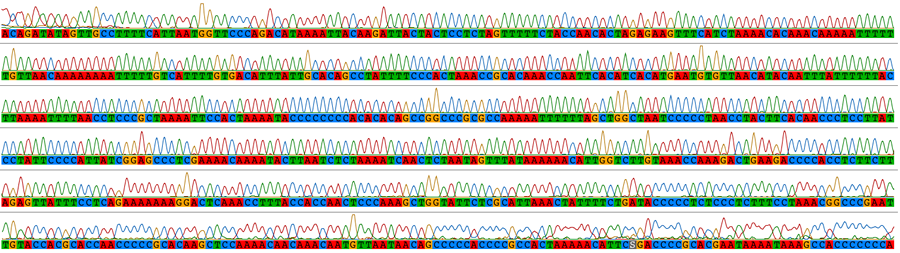
Traditional sequencing allows the targeted amplification of particular genes of interest. DNA is amplified by polymerase chain reaction (PCR) using specific primers and sequenced using Sanger sequencing. The resulting electropherograms (below) are checked for errors then aligned (bottom) for phylogenetic analysis.
PCR is powerful because it can make use of small amounts of starting DNA which means that even old specimens and some fossils can provide a source DNA. For this we use a dedicated ancient DNA lab (see aDNA). PCR also allows data to be collected from rare or small species and as little tissue as a single hair or feather tip can be sufficient. This is especially valuable for studies of conservation genetics. PCR allows particular genes or parts of a genome to be targeted so the resulting data can be readily aligned for comparison. PCR is also used to target microsatellites (short repeat sequences) for genotyping.
In our lab, PCR has been used to generate data in a wide range of population genetics, phylogeographic and phylogenetic studies of birds, lizards, insects and other animals. Below is an alignment of mitochondrial sequence from species of rail.

Next Gen sequencing has revolutionized the way we collect data for molecular ecology and evolution. Millions of short sequence reads are produced in parallel, and must be assembled into longer contigs using mapping or de-novo sequence assembly methods.
Among the applications of these approaches employed in our lab are the generation of entire mitochondrial DNA genome sequences of birds (e.g. studies of speciation in rails, Rallidae), sequencing of nuclear genes of insects using copyDNA from messengerRNA extracted from target species, and compilation of large scale population genetic
arrays using tools such as restriction site associated markers.
Karyotypes, Cytology & Flow-cytometry
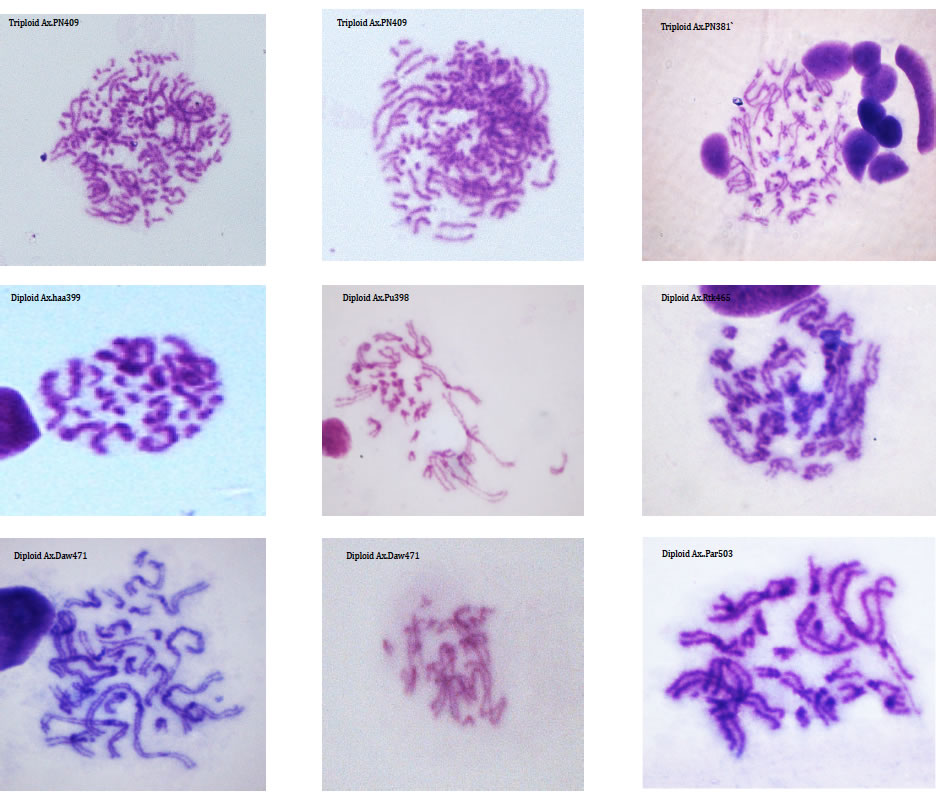
DNA is organised into chromosomes that, during cell division, can be seen and studied using light microscopes. By studying the number of chromosomes within individuals, populations and species we can document genetic variation. The relative size and centromere position of chromosomes is used to describe the complete set of chromosomes (the karyotype) of a taxon.
Although many species have a characteristic genome size and karyotypes, there is a good deal of variation within some lineages. For example the Auckland tree weta, Hemideina thoracica, has at least eight different chromosome races, each with its own distinctive karyotype (Morgan-Richards 1997). Intraspecific karyotype variation is not concordant with allozyme variation in the Auckland tree weta of New Zealand. We use plain staining of chromosomes from insects (using Geimsa) to document chromosome rearrangements and study hybrid zones (Morgan-Richards et al. 2001) and understand changes in ploidy (Myers et al. 2012).
Ploidy level variation can also be investigated by measuring the amount of DNA per cell. Estimates of genome size using flow-cytometry are important for understanding chromosome rearrangements (Morgan-Richards 2005) and can aid identification of hybrids (Morgan-Richards et al. 2004). Increasingly, genome size estimates are important during high-throughput DNA sequencing projects.
Data Files from various studies can be downloaded from here:
| PUBLICATION |
click on icon |
Koot EM, Morgan-Richards M,Trewick SA. Climate change and alpine-adapted insects: modelling environmental envelopes of a grasshopper radiation.
|
 |
Pattabiraman N, Morgan-RIchards M, Powlesland R, Trewick SA. Unrestricted gene flow between two subspecies of translocated brushtail possums (Trichosurus vulpecula) in New Zealand.
|
 |
Morgan-Richards M, Vilcot M, Trewick SA. Lack of assortative mating in a modified landscape leads to a hybrid swarm.
|
 |
Carmelet D, Morgan-Richards M, Koot EM, Trewick SA. Climate and ice in the last glacial maximum explain patterns of isolation by5 distance inferred for alpine grasshoppers.
|
 |
Daly EE, Trewick SA, Dowle EJ, Crampton JS, Morgan-Richards M. Conservation of pūpū whakarongotaua - the snail that listens for the war party.
|
 |
Quenu M, Trewick SA, Bresica F, Morgan-Richards M. Association of machine learning algorithms and geometric morphometric suggest hybridisation of two land snail species in the Isle of Pines.
|
 |
Morgan-Richards M, Langton-Myers SS, Trewick SA. Labile reproductive method of a stick insect species suggests short-term advantages for sex.
|
 |
Bridgeman B, Morgan-Richards M, Wheeler D, Trewicks SA. First detection of Wolbachia in the New Zealand biota.
|
 |
Morgan-Richard M, Bulgarella M, Sivyer L, Dowle EJ, Hale M, van Heugten R, McKean NE, Trewick SA. 2017. Explaining large mitochondrial sequence differences within a population sample.
|
 |
Bradley E, Trewick SA, Morgan-Richards M. 2017 Distinctiveness of a mouse population suggests low gene flow between Waikawa Island and mainland New Zealand. New Zealand Journal of Ecology.
|
 |
McKean NE, Trewick SA, Morgan-Richards M. 2016. Little or no gene flow despite F1 hybrids at two interspecific contact zones. Ecology and Evolution.
|
 |
Morgan-Richards M, Hills SFK, Biggs P, Trewick SA. 2016 Sticky genomes: using NGS evidence to test hybrid speciation hypothesis. PLOS One.
|
 |
Nelson-Tunley M, Morgan-Richards M, Trewick SA. 2016. Genetic diversity and gene flow in a rare New Zealand skink despite fragmented habitat in a volcanic landscape. Biological Journal of the Linnean Society.
|
 |
|